Rational Drug Design
7 min read
Prof. H. S. Savithri
NASI Senior Scientist,
Department of Biochemistry,
Indian Institute of Science, Bengaluru 560012
“When I woke up just after dawn on September 28, 1928, I certainly didn’t plan to revolutionize all medicine by discovering the world’s first antibiotic, or bacteria killer”
– Alexander Fleming
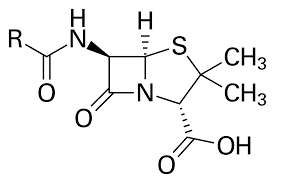
Alexander Fleming discovered Penicillin (Fig. 1) and even today its derivatives are the drug of choice for many bacterial diseases. Indeed this discovery revolutionized medicine. He was awarded the Nobel Prize in 1945 along with Howard Florey and Ernst Chain, who isolated the pure compound, Penicillin F, from the fungus Penicillium rubens. Penicillin and its derivatives (also called β-lactam antibiotics) are now synthesized chemically and are effective against many bacterial infections, in particular those caused by staphylococci and streptococci. This discovery of penicillin was serendipitous. Later extensive research lead to the elucidation of its mechanism of action and also the mechanism by which the bacterial pathogens develop resistance. Penicillin kills bacteria by inhibiting the activity of transpeptidases (also called penicillin binding proteins) that are needed for the cross-linking of peptidoglycans in the final step in bacterial cell wall biosynthesis. The fewer cross-links weaken the cell wall leading to uncontrollable flow of water into the bacterial cells. This results in cell lysis and death. Some bacteria produce beta lactamases, which can cleave the beta lactum ring of penicillin that makes them resistant to penicillin. In such cases, derivatives of penicillin or a combination with drugs that inhibit the lactamases are used.
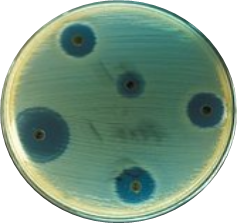
Subsequent to the work of Fleming, Selman Waksman devised the zone of clearance technique for identifying natural substances that exhibit antimicrobial properties. Initially a bacterial culture is plated on an agar plate and allowed to grow over night. Then, small holes are made and different compounds/extracts are put in them and they are allowed to diffuse into the agar. If the extract has antimicrobial properties, it will kill the cells leading to a zone of clearance. The diameter of the zone cleared is an indication of the relative effectiveness of the extract (Fig.2). Even today this technique is widely used for the screening of various extracts.
Selman Walksman was awarded the Nobel Prize in physiology or medicine in 1952 “for his discovery of streptomycin, the first antibiotic effective against tuberculosis. He is considered the father of antibiotics.
Discovery of a new drug involves many steps: target identification, validation, lead compound identification, lead optimization followed by preclinical and clinical trials. This generally takes tens of years with several scientists working together. More recently there have been attempts to reduce this time by a adopting a rational approach to the design and development of new drugs. These improvements are discussed in the following sections.
Target identification
The identification of new, clinically relevant, molecular targets is of utmost importance for the discovery of innovative drugs. If we assume that there may be approximately 10 genes involved in a particular disease with some 5 to 10 additional genes linked to the disease and that there are 100 major diseases, then there could be 5000 to 10,000 targets that could be used for drug design.
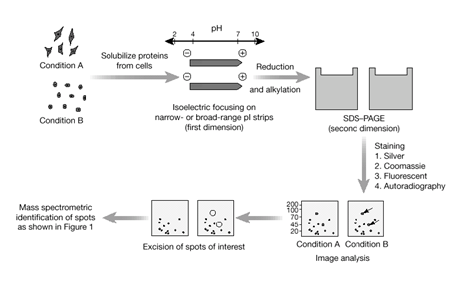
However, only 500 unique targets are being used currently for therapy. Thus, many more drug targets exist and the challenge is to identify such novel targets. A drug target is a molecule involved in a particular metabolic or signaling pathway that is specific to a disease condition or pathology or to the infectivity or survival of a pathogen. As mentioned earlier, initially such targets were discovered serendipitously. They were also identified following traditional folklore remedies. However, in the recent years, a thorough analysis of the molecular and physiological processes of a disease is done to identify the most probable targets. Complete genome sequence of organisms causing the disease, the expression profile of their genes (referred to as tanscriptomes), their protein profile (proteome) are first determined. There are methods that can be used to compare the protein profile of a diseased person with that of a normal person or compare the protein profile of a normal cell with that of a cancer cell or between the host and the pathogen. Such an analysis is referred to as proteome analysis. In this method thousands of proteins from a cell/tissue extract are separated by what is called as 2D electrophoresis (Fig. 3). The proteins are separated in one dimension (one direction of the separating gel) on the basis of their charge and in the other dimension (a perpendicular direction in the separating gel) on the basis of their size. The resolution of such 2D gels has improved enormously and proteins with very small charge and size differences can be separated and visualized as distinct spots. Proteins unique to the disease can thus be identified by computer aided comparison of the maps.
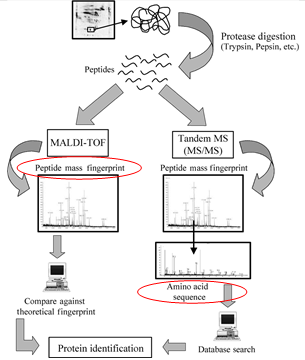
The proteins present in picomolar quantities in the unique spots are identified by mass spectrometric analysis. The present day mass spectrometers can analyze and determine the molecular mass of proteins with an accuracy 0.005-0.1%. By such an accurate analysis, not only of the mass of the protein but also the peptides derived from it by enzymatic cleavage can be deciphered. (Fig. 4). A comparison against the theoretical finger print (mass of cleaved fragments) of all unique proteins in the protein sequence data base helps to identify the target proteins.
Target Validation
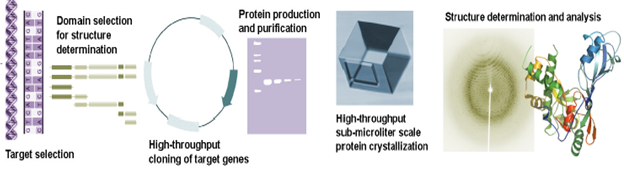
The targets identified for a particular disease need to be validated. This is done in several steps. A systematic analysis of the target protein sequence with those present in the data base is carried out to detect if there are any mutations that could be linked with the disease. The expression level of the target protein is checked to assess if it has changed during the course of the disease. One may also analyze if the target protein is structurally similar to proteins that have been previously identified as reasonably good drug targets. In order to gain further insights into the target protein, the corresponding gene segment is amplified by polymerase chain reaction (PCR), over-expressed using appropriate vectors in heterologous systems, purified, crystallized and the three dimensional structure of the protein is determined (Fig. 5). There has been such a major technological revolution in these methods that the time scale has reduced from several years to a couple of months.
Lead compound identification
The structure of the target protein is analyzed using bioinformatic tools to identify probable ligand/drug binding pockets, molecules that can bind and possibly inhibit the function of the target protein. The affinity of the ligand for the target protein could be enhanced by. Computer aided design of molecules using various docking and modeling programs. A theoretical analysis of the binding constants of these molecules in silico is carried out to select molecules which have a very low Ki value, such compounds are likely to inhibit the target protein function. The compounds are then synthesized and tested for their ability to inhibit the function of the target protein in vitro. Further changes in the functional groups are made to improve solubility/ strength of binding etc and the newly synthesized compounds are once again tested for their ability to inhibit the function of the target protein. The most promising candidates are tested for their effect on the in vivo function of the target protein. Expert scientists from different fields such as Biochemistry, Molecular Biology, Cell Biology Organic Chemistry, Bioinformatics and Structural Biology work together to find promising leads.
Lead compound optimization
Methods have to be developed for large scale synthesis, which could reduce the cost of the drug, and the designed drugs should have lowest Ki of target protein-inhibitor complex and improved potency. Further changes to the molecule are made such that it would have reduced off-target activities (leading to fewer side effects). The physicochemical and metabolic properties of the molecules are also analysed in detail. Sometimes the accumulation of the substrates of a reaction inhibited can lead to what is known as metabolic resistance. For example, a compound called phophonoacetyl L aspartate, (PAcAsp), a tight binding inhibitor of aspartate transcarbamylase, is ineffective in vivo as it results in the accumulation of carbomyl phosphate, one of the substrates of the reaction catalyzed by this enzyme. Lead optimization is also required for the compounds isolated from natural sources. For example, Artemisinin, a leading drug in the treatment for malaria, is extracted from a grass that is found in China and Vietnam. However, it has poor solubility and hence is less absorbed into the blood stream. Further the compound is present in very low amounts in the natural source. Efforts to improve the solubility and absorption of artemisinin have lead to the manufacture of compound with increased yield and better absorption.
Preclinical and clinical development
The identified potential molecule is initially tested in experimental animal models (preclinical testing) to asses if it exhibits. drug like properties. The questions that are addressed are – Is it specific to the target? Is the target protein specific to only one isomorph of the compound? What are its ADME-Absorption, Distribution, Metabolism, Excretion properties? The liver is the organ that metabolizes most of the molecules such that they are eventually excreted in the urine. If the molecule is excreted before it can reach the site of action, the drug is likely to be ineffective. The bioavailability, different delivery methods, routes administration, dosage and their regimen need to be optimized. Toxicity, cross reactions, side effects also are assessed thoroughly before the drug is ready for clinical trials. Clinical trials are conducted with approval from concerned authorities, informed consent from the subjects, in appropriately chosen locations using double blind methods. The data obtained are analyzed by experts before the drug is approved for release into the market. Experts in the field of Pharmacology, Medicine, Statistics and Business Administration work together to achieve this. This rational drug design is truly a multidisciplinary effort that needs expertise from a large number of disciplines.






